

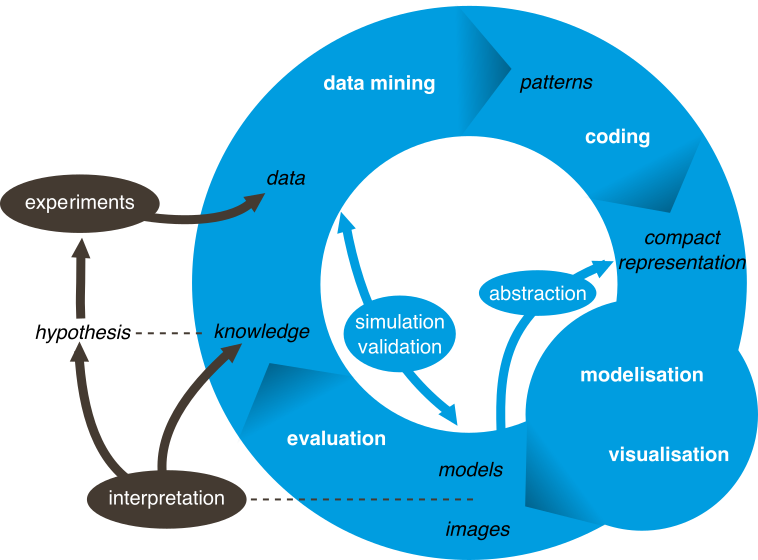
The team MABioVis develops a scientific vision inspired by the synergy now taking place between information sciences, life sciences, social sciences and economics. The combination of experimental techniques for high-speed and advanced computational methods allows to tackle problems of unprecedented size, and gives birth to scientific issues emerging in a variety of fields such as biology and quantitative geography.
In the different areas that we study, the data can be modeled by complex systems defined as sets of interacting entities. Members of the MABioVis team contribute to the development and study of formalisms, models and algorithms providing results on both extraction and data management, but also on the construction, analysis and understanding of these complex systems. The results obtained and the ties with academic and industrial partners show that we have been successful at addressing this challenge in biology (life systems) and quantitative geography (urban networks).
Several members of our group are also a part of the PLEIADE and
MNEMOSYNE projects from INRIA.
Some members of our group are involved in the management of the
Our team is actively involved in the rapid development of Synthetic Biology activities in Bordeaux (SB2).
The research conducted in CoBalt takes its source in many areas in biology or bio-informatics and from biological data: genomic and protein sequences, RNA, biological interaction networks, metabolic and signaling pathways. We aim at the development of new algorithms and formal models for the analysis of genomes, networks, and more generally the study of complex systems. We ultimately support biologists in their understanding of the structure and the history of genomes. We thus address challenges initiated from problems in bioinformatics while encompassing issues in computer science: algorithmic recognition and inference patterns, data mining and classification, modeling of complex dynamical systems, prediction and comparison of networks, comparative genomics in a broad sense. Full details
The research conducted in EVADoMe is based on three main lines of research: data mining, graph drawing and visual interactive exploration of data. Data mining can be viewed as an upstream process for visual data analysis. Graph drawing consists in emebedding the vertices of a graph in a geometrical space (a 2D Euclidean space is commonly used in information visualization). When designing a visualization system, often starting from user tasks and available data, one must decide of a proper visual representation coupled with task-relevant interactions. We have investigated the use of GPU’s to improve interactivity when exploring sophisticated graph drawing for large graphs (typically using curves for edges). Full details

Sofian Maabout
Sofian [dot] Maabout [at] labri [dot] fr
Phone: +33 540 003 527

Mathieu Raffinot
Mathieu [dot] Raffinot [at] labri [dot] fr
Phone: +33 540 003 610